Biologie structurale, Modélisation des Interactions Moléculaires, Ingénierie Cristalline
L’équipe BioMIMIC réunit des chercheurs aux compétences et expertises variées, allant de la physique à la biologie, qui se consacrent à la compréhension approfondie des systèmes moléculaires à l’échelle atomique. Grâce à cette diversité de connaissances et à leur objectif commun, ils sont en mesure de mener des recherches de pointe dans les domaines de la Biologie Structurale, de la Modélisation des Interactions Moléculaires et de l’Ingénierie Cristalline.

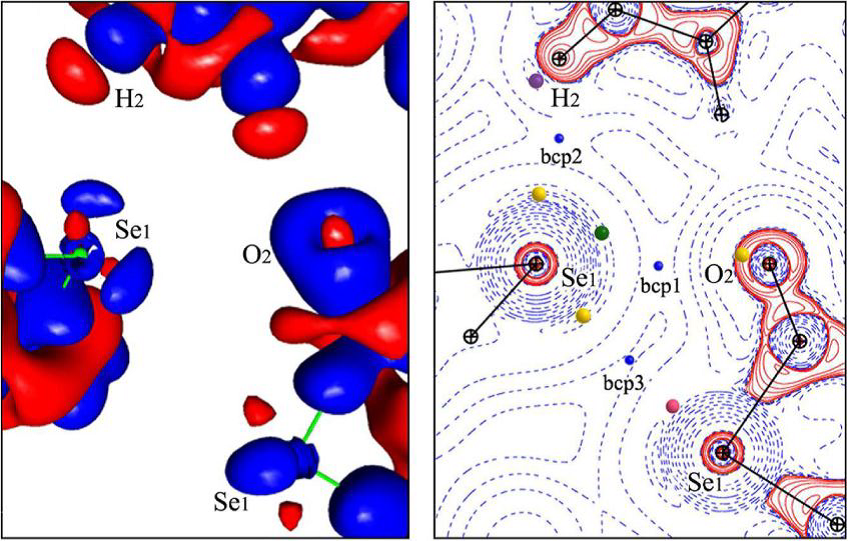
Dans ces domaines, des méthodes originales et reconnues internationalement sont développées visant à exploiter la densité de charge issue de calculs de chimie quantique ou multipolaire expérimentale déduite de la cristallographie à ultra-haute résolution. La détermination correcte de ces propriétés est fondamentale puisque les forces électrostatiques sont à l’origine de la reconnaissance moléculaire et des réactions biochimiques. L’examen de la topologie de la densité électronique et de son Laplacien permet de caractériser les interactions intermoléculaires, tant du point de vue de leur orientation que de leur amplitude. Ces développements offrent des outils pertinents pour analyser les interactions telles que l’interaction trou-sigma qui est directement liée à la réactivité moléculaire. En outre, des méthodes éprouvées, telles que la RMN, la diffraction des rayons X et la modélisation in silico, sont utilisées pour étudier les relations structure-fonction d’enzymes mais aussi de protéines structurales.
Responsable(s)
- DIDIERJEAN Claude, Professeur des universités
- JELSCH Christian, Directeur de recherche CNRS
Membres
- AUBERT Emmanuel, Maître de conférences
- BEGUE Dunkan, Doctorant
- ESPINOSA Enrique, Directeur du laboratoire et Professeur des universités
- FAVIER Frédérique, Maître de conférences
- FRANCOIS Romain, ATER
- GUILLOT Benoît, Professeur des universités
- LECOMTE Claude, Professeur émérite
- MAMBATTA Haritha, Post-Doctorant
- MATHIOT Sandrine, Technicien
- SEKKOURI ALAOUI Hicham, Post-Doctorant
- TSAN Pascale, Maître de conférences
Structure-function relationships
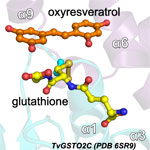
We are recognized for our ability to experimentally obtain three-dimensional structures of biological macromolecules and to analyze their structure function relationships. A first research topic on which we have more than 10 years of expertise concerns glutathione transferases and their complexes with small ligands.
A second one is focused on the characterization, at the molecular level and from a fundamental point of view, of the bacterial conjugation allowing these microorganisms to evolve rapidly, for example in order to resist an antibiotic.
We also have an expertise in the analysis of the conformations of peptidomimetics in the solid state.
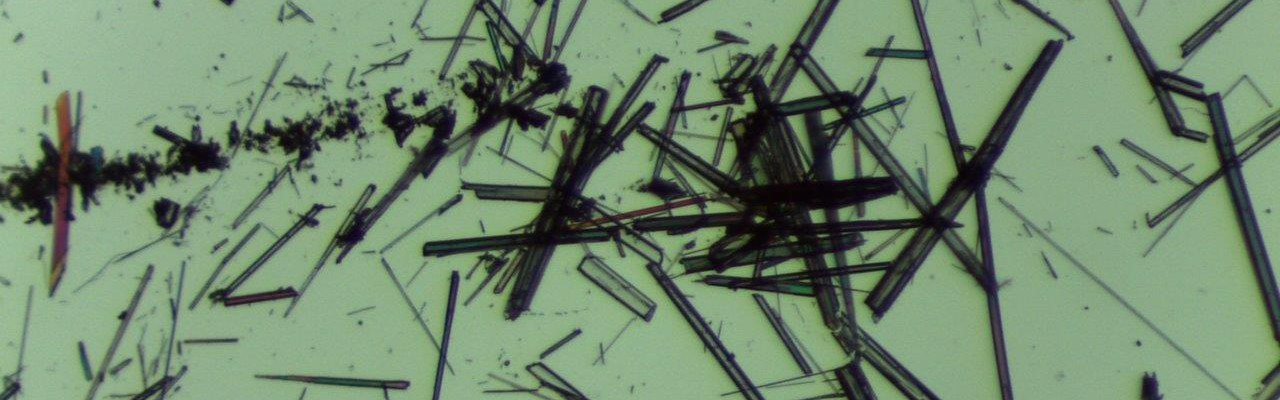
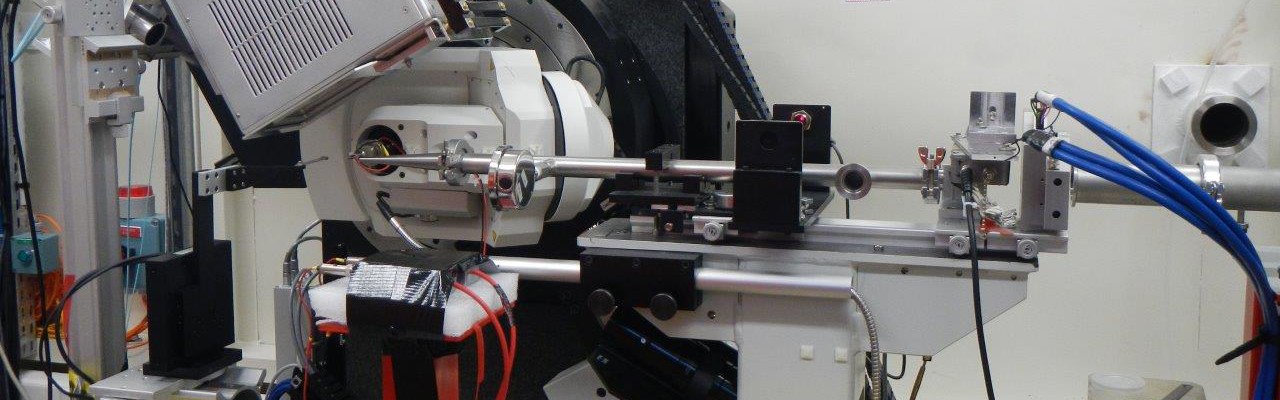
We determine the structures by X-ray crystallography or by NMR. Our crystallogenesis platform includes two pipetting robots and a semi-automatic binocular. It allows us to perform many tests and increases the chances of obtaining crystals. Thanks to the X-ray diffraction and scattering platform hosted by the laboratory, we can perform preliminary tests of the crystals at home, and even collect experimental data necessary to establish a preliminary structure. Our access to the SOLEIL and ESRF synchrotrons allows us to improve them at maximum resolution.
Molecular modelling at ultra high resolution
An extensive survey of Cambridge Structural Database is carried out to study directionality and stereochemistry of hydrogen bonds with an oxygen acceptor including carbonyl, alcohols, phenols, ethers and esters groups. The results obtained through this survey are correlated with the charge density of these different chemical groups. The electron density of these different oxygen atoms types show striking dissimilarities in the electron lone pairs configuration.
Crystal Enginering
The Crystal Engineering group focuses on the control of both molecular organization in space and modulation of the intensity of the intermolecular interactions involved in this organization. The study of directional intermolecular interactions is at the center of our approach. It relies on the use of the influence of intra- and intermolecular environment in order to drive the crystalline molecular arrangement, and thus to modify the physicochemical properties of the crystalline solids. The study of the electron distribution and its derived properties from experimental (XRD) and theoretical (quantum calculations) methods allows us to characterize the interatomic and intermolecular interactions involved on.
Software MoProSuite
The refinement program MoPro (Molecular Properties) is dedicated to the refinement at (sub)atomic resolution of structures ranging from small molecules to biological macromolecules. MoPro uses the multipolar pseudo-atom model for the electron-density refinement. Alternative methods are also proposed, such as modelling bonding and lone-pair electron density by virtual spherical atoms.