Summary
Bacterial genomes adapt to environmental changes through rapid evolutionary mechanisms, namely the acquisition of genes by horizontal transfer. These genes allow them to gain properties such as antibiotic resistance. The transferred genes are present in mobile elements such as integrative and conjugative elements (ICEs) included in the bacterial chromosome. They encode their own excision, transfer and integration, in addition to cargo genes that provide advantageous properties to the host. The whole conjugation machinery is very complex and involves many proteins. It remains poorly understood, especially in Gram-positive bacteria. The model chosen in our study is ICESt3 from Streptococcus thermophilus. To initiate this work, the genes coding for the OrfA, G, J, L and M proteins were selected. We solved the crystallographic structure of the soluble domain of OrfG, homologous to VirB8, a central protein of the type IV secretion system (T4SS) of Gram-negative bacteria. The smaller OrfL and M proteins are studied by NMR in our team. Our ambitious goal is to study complex macromolecular assemblies involved in conjugation, with obvious potential applications in terms of public health.
Collaborations: Pr. N. Leblond (DynAMic, UMR UL-INRA 1128, Nancy).
Bacterial genomes adapt to environmental changes throughbiomod_fig_2 rapid evolutionary mechanisms, namely the acquisition of genes by horizontal transfer. These genes allow them to gain properties such as antibiotic resistance which remains a major public health problem. The transferred genes are present in mobile elements such as integrative and conjugative elements (ICEs) included in the bacterial chromosome. They encode their own excision, transfer and integration, in addition to cargo genes that provide advantageous properties to the host. The whole conjugation machinery is very complex and involves many proteins. It remains poorly understood, especially in Gram-positive bacteria. Our collaborators of the DynAMic laboratory (ICE-TeA team of the UMR UL-INRA 1128, Nancy) are well known for their expertise on Gram-positive bacterial ICEs. Their model is an ICE from Streptococcus thermophilus, ICESt3.
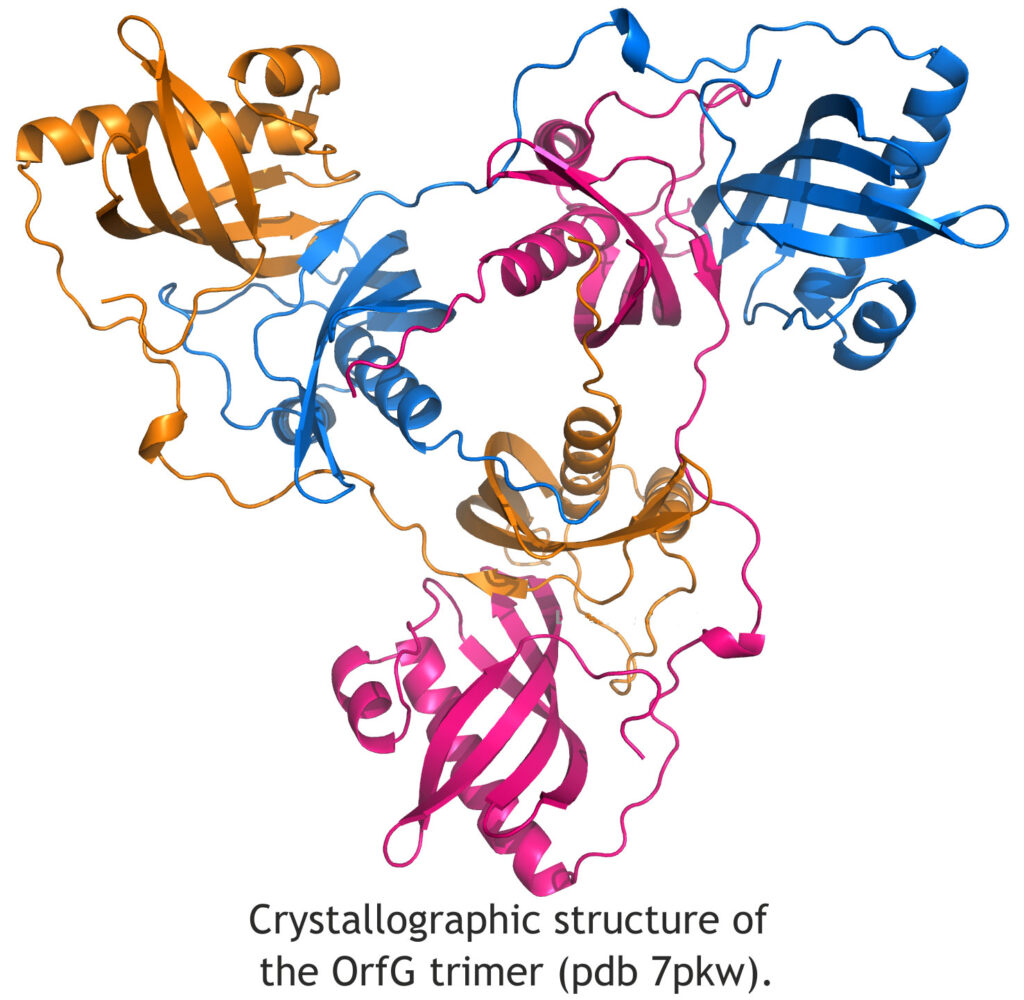
Several genes were targeted to initiate this study, both genes encoding proteins with identified homologues in Gram-negative bacteria (OrfA, OrfG and OrfJ) but also genes encoding ICE-specific proteins (OrfL and OrfM). In this project, our team is involved in the structural characterization of these proteins. After many difficulties in crystallogenesis and in the exploitation of experimental data, we succeeded in solving the crystallographic structure of the soluble domain of OrfG (i). OrfG is homologous to VirB8 which is a central protein of the type IV secretion system (T4SS) in Agrobacterium tumefaciens (Gram-negative). Despite very low sequence identity, these two proteins have comparable tertiary structures. However, their assembly properties in the conjugation pore are certainly very different, and we obtained a trimeric quaternary structure that is different from those usually observed in Gram-negative bacteria. The original entanglement of the subunits within this trimer raises questions about the representativeness of this unit in vivo (figure), and studies are underway to investigate its probability.
In the longer term, this project aims at the ambitious study of very complex macromolecular assemblies whose three-dimensional structures will clarify our knowledge on the type IV secretion systems of Gram-positive bacteria. This study is important from a fundamental point of view because studies show that the partially known Gram-negative model cannot be transferred to Gram-positive model. Clearly, understanding the processes of bacterial gene acquisition has the potential to generate public health applications.
References:
- “Structural and Biochemical Analysis of OrfG: The VirB8-like Component of the Conjugative Type IV Secretion System of ICESt3 From Streptococcus thermophilus”, Cappele, Mohamad-Ali, Leblond-Bourget, Mathiot, Dhalleine, Payot, Savko, Didierjean, Favier, Douzi, Front. Mol. Biosci. (2021) 8:642606. DOI: https://doi.org/10.3389/fmolb.2021.642606 HAL: hal-03196531v1.